#NMR observation of methyl groups in proteins from eukaryotic and cell-free expression systems
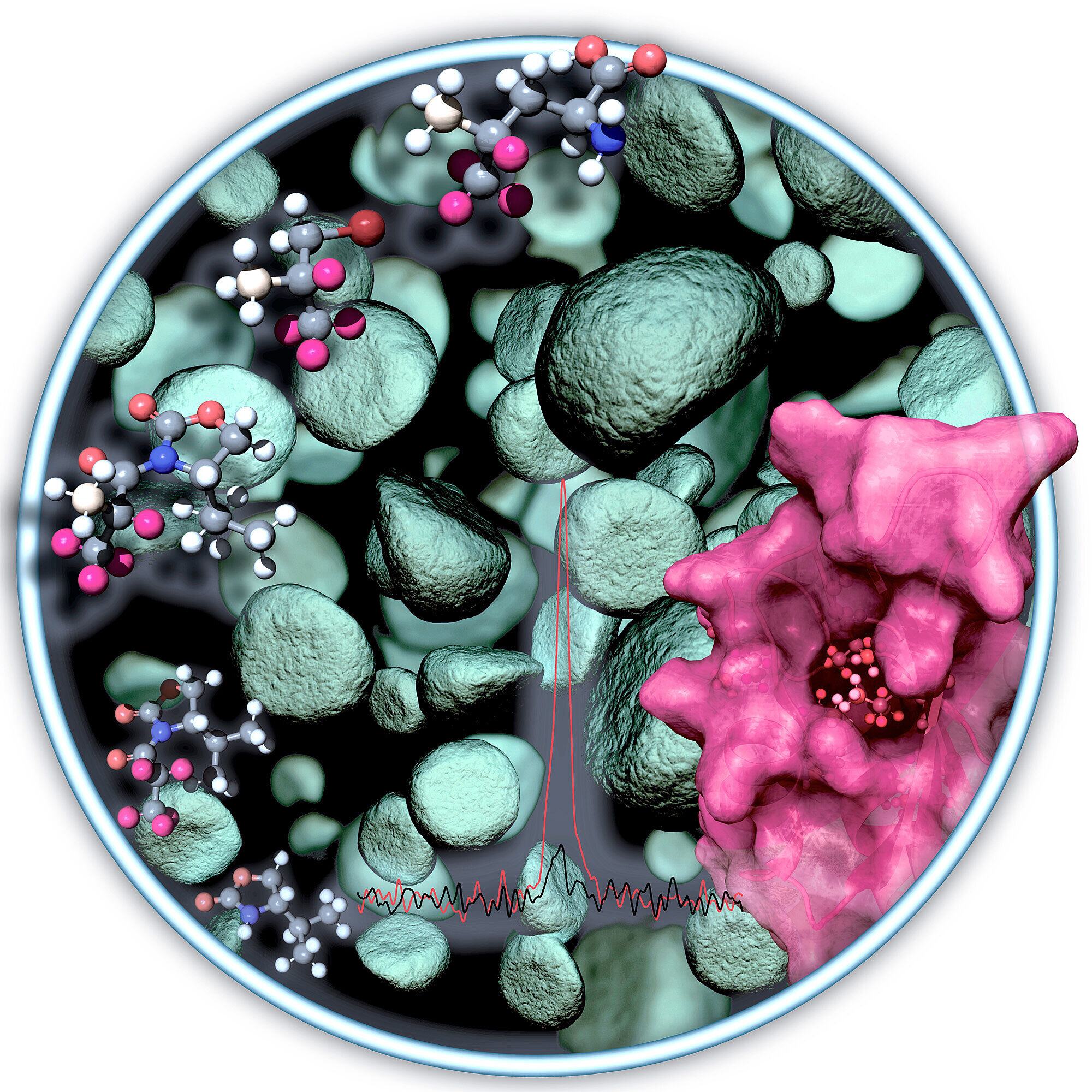
“#NMR observation of methyl groups in proteins from eukaryotic and cell-free expression systems”
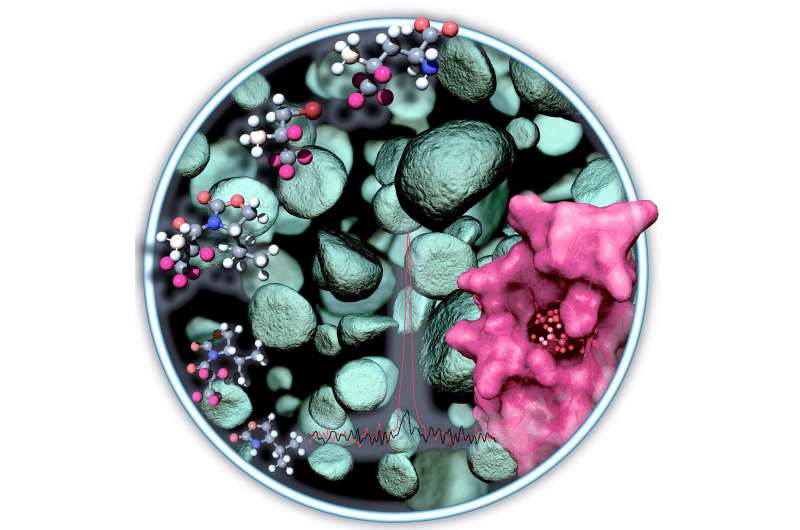
An international research team involving the working group of biophysicist Dr. Manuel Etzkorn from Heinrich Heine University Düsseldorf (HHU) has developed an approach for using NMR spectrometry to analyze important molecules that have not been accessible before now. In the journal Angewandte Chemie, the authors describe a simpler and efficient means of labeling the molecules with methyl groups as sensors.
To understand life at molecular level, we need to be able to investigate the central components such as proteins in as natural a form and environment as possible. NMR or ‘nuclear magnetic resonance’ spectroscopy offers unique opportunities in this regard.
Methyl groups within proteins are particularly suitable sensors for use in this method. Methyl groups are composed of one carbon and three hydrogen atoms. To amplify the signal from these sensors to a sufficient level, large portions of the remaining protein need to be enriched with deuterium atoms in complicated procedures. Deuterium is a hydrogen isotope with a proton and a neutron in its atomic nucleus. In the past, however, such enrichment was possible only using special production platforms.
As a result, the use of NMR spectroscopy to investigate systems that could not be produced using those platforms was either impossible or very limited. Such systems include in particular a whole range of therapeutically relevant systems such as antibodies or the class of GPCRs (“G-protein coupled receptors’) targeted by a very large percentage of modern medicines.
A research team at HHU led by Dr. Manuel Etzkorn from the Institute of Physical Biology and the Biomolecular NMR Center (which is run jointly by HHU and Forschungszentrum Jülich) collaborated with colleagues from Sofia University, Harvard Medical School and the Dana Faber Cancer Institute in Boston to develop a new method that can integrate the characteristics required of the sensors in all common production platforms. The synthesis method is considerably simpler and over 20 times more cost effective than previous methods for integrating methyl group sensors. It also works in systems that were inaccessible up to now.
Angewandte Chemie put the research work on the front cover of its current edition. This highlights the special importance of this topic for improving biophysical basic research and the resulting development of new kinds of medicines.
Dr. Etzkorn says, “The new method will allow us and others to investigate the building blocks of life in an amazing level of detail and in environments that are as natural as possible.”
Cell growth: Intricate network of potential new regulatory mechanisms has been decoded
Abhinav Dubey et al. Local Deuteration Enables NMR Observation of Methyl Groups in Proteins from Eukaryotic and Cell‐Free Expression Systems, Angewandte Chemie International Edition (2021). DOI: 10.1002/anie.202016070
Provided by
Heinrich-Heine University Duesseldorf
Citation:
NMR observation of methyl groups in proteins from eukaryotic and cell-free expression systems (2021, May 17)
retrieved 17 May 2021
from https://phys.org/news/2021-05-nmr-methyl-groups-proteins-eukaryotic.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
If you liked the article, do not forget to share it with your friends. Follow us on Google News too, click on the star and choose us from your favorites.
For forums sites go to Forum.BuradaBiliyorum.Com
If you want to read more Like this articles, you can visit our Science category.