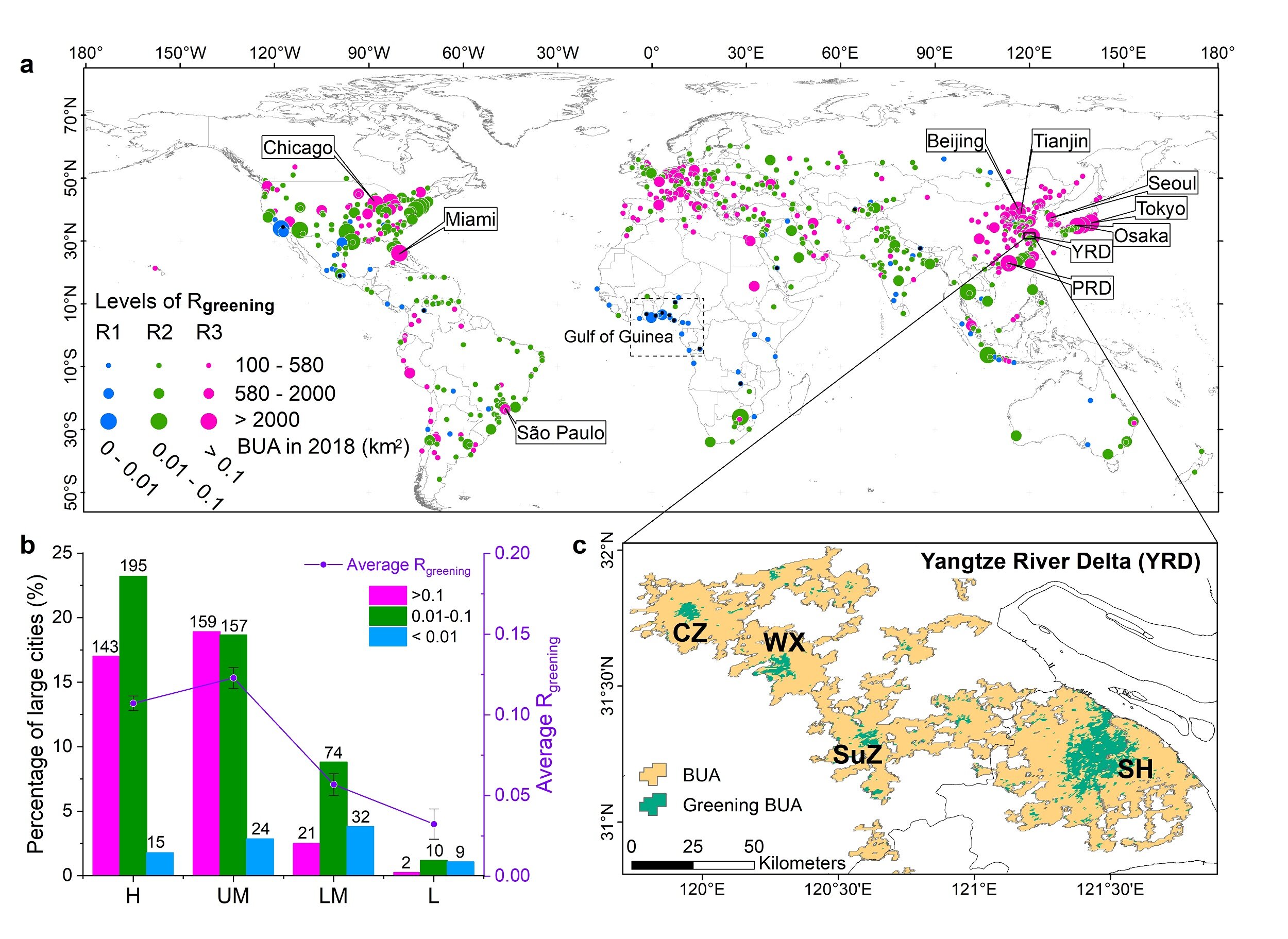
#The modeling and simulation of self-organized intracellular twisters in the Drosophila oocyte


Cytoplasmic streaming is the large-scale motion of cytoplasm (i.e., gelatinous liquid inside cells) inside a living cell. This flow, known to regulate various intracellular processes, can vary greatly between different cell types at different stages of a cell’s development. Examining and modeling the different types of cytoplasmic flows can help us understand how they emerge in specific types of cells.
Past studies primarily examined streaming cytoplasmic flows in large cells where, it is often argued, diffusion is too slow to enable biological processes that organisms need to perform (e.g., development of an egg or of an embryo, or in large plant cells).
As a result of this slow diffusion, the flow enables a faster distribution of cellular components. In early fly oocytes (i.e., developing egg cells), for instance, cytoplasmic streaming appears random, while at later stages of development, where the oocyte is larger, they can appear large-scale and rotational.
Researchers at the Flatiron Institute, building upon previous work, recently introduced a versatile modeling strategy that can be used to study self-organized cytoplasmic streaming in systems comprised of hydrodynamically coupled deformable fibers.
This model, introduced in Nature Physics and in collaboration with scientists at Princeton and Northwestern universities, was combined with data collected in experiments on the Drosophila (i.e., fruit fly) oocyte to gather insights about self-organized cytoplasmic flow.
“I’ve been working in the general areas of biologically active matter, intracellular mechanics, and complex fluids for a while,” Michael J. Shelley, co-author of the paper, told Phys.org. “The problem tackled in our recent paper combines all these areas, each of which I really like.
“I learned about this particular problem of flows in oocytes from my friend Ray Goldstein and realized that earlier work with my Flatiron colleague David Stein might be adapted to understand something about the oocyte problem. It did, and David and I worked together with Ray and his colleagues at Cambridge on a first very stripped down 2D model.” That work was published in Physical Review Letters in 2021.
Researchers at the Flatiron Institute had previously developed various tools to study the hydrodynamics of moving microtubules, stiff biopolymers that are a central element of the cell’s cytoskeleton. Shelley, Stein and their colleagues Reza Farhadifar, Sayantan Dutta, and Stas Shvartsman planned to use these numerical tools to study the onset of self-organized cytoplasmic flows in 3D cells.
“The primary objective of our recent study was to provide a minimal, but not too minimal, model invoking only microtubules, molecular motors, and cytoplasm that could explain experimental observations and help to make predictions,” Shelley explained.
The recent study carried out by Shelley and his colleagues combines physics and mathematical theories with experimental results. The researchers started by creating a model that they could then use to simulate self-organized cytoplasmic streaming in the Drosophila oocyte.
“We wrote down a mathematical model for the stresses that molecular motors create by moving on a microtubule,” Shelley said. “This model should allow the microtubule to bend under loads, and for its bending to move cytoplasm which affects the bending of other microtubules. Next, used a high-quality piece of software—here called SkellySim—which lets you simulate a few thousand of such microtubules interacting by collectively pushing fluid as they collectively bend.”
After developing their model and running simulations, Shelley and his colleagues carried out experiments on Drosophila oocytes. Firstly, they used light microscopy to examine cytoplasmic motions in developing egg cells and then analyzed the data they collected using particle imaging velocimetry to reconstruct cytoplasmic velocity fields.
“Our paper provides a clear example of how, with a very few ingredients, of how a large-scale transport system (i.e., the streaming flow) could emerge in the cell from the interactions of just a few components (i.e., microtubules, motors and cytoplasm),” Shelley said. “The beauty lies in its robustness, as in large parts of the parameter space controlling the model the system just wants to form a twister. This is a great example, I think, of biological self-organization to perform a task.”
Notably, using their model, the researchers were also able to predict the effect of cell shape on the orientation of twisters. Their predictions suggest that while in the dynamics of cytoplasmic streaming in Drosophila oocytes could be incredibly complex, they ultimately result in a simple final state (i.e., a twister).
The findings gathered by Shelley and his collaborators could soon pave the way for further explorations of cytoplasmic streaming, specifically focusing on this simple twister state. This could lead to interesting new discoveries about the physics underpinning vital processes in biological cells.
“This work demonstrated the power that high performance computing and modern algorithms can bring to understanding biophysical phenomena,” Shelley added. “In our next studies, we plan to explore how these twister flows mix components across the cell or enable their delivery from one point to another.
“There are other transport systems within oocytes, like through ring canals, that are very interesting. I am generally interested in the manifold ways that the cellular cytoskeleton organizes itself in order to get cellular things done.”
Sayantan Dutta et al, Self-organized intracellular twisters, Nature Physics (2024). DOI: 10.1038/s41567-023-02372-1
© 2024 Science X Network
Citation:
The modeling and simulation of self-organized intracellular twisters in the Drosophila oocyte (2024, February 21)
retrieved 21 February 2024
from https://phys.org/news/2024-02-simulation-intracellular-twisters-drosophila-oocyte.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
If you liked the article, do not forget to share it with your friends. Follow us on Google News too, click on the star and choose us from your favorites.
If you want to read more Like this articles, you can visit our Science category.